Nonparametric (model-free) estimators of causal effects
This page illustrates nonparametric adjustment: how to estimate causal effects without any statistical model at all. Although it is rarely used in practice, nonparametric adjustment is a conceptually useful because it illustrates the ideal case that we hope any statistical model will approximate.
There are two steps:
1) Estimate effects within population subgroups defined by confounders
Why? Within subgroups, confounders do not vary. Differences in outcomes must be caused by the treatment!
2) Aggregate over subgroups, weighted by size
Why? The distribution of confounders stays the same, even if we intervene to change the treatment
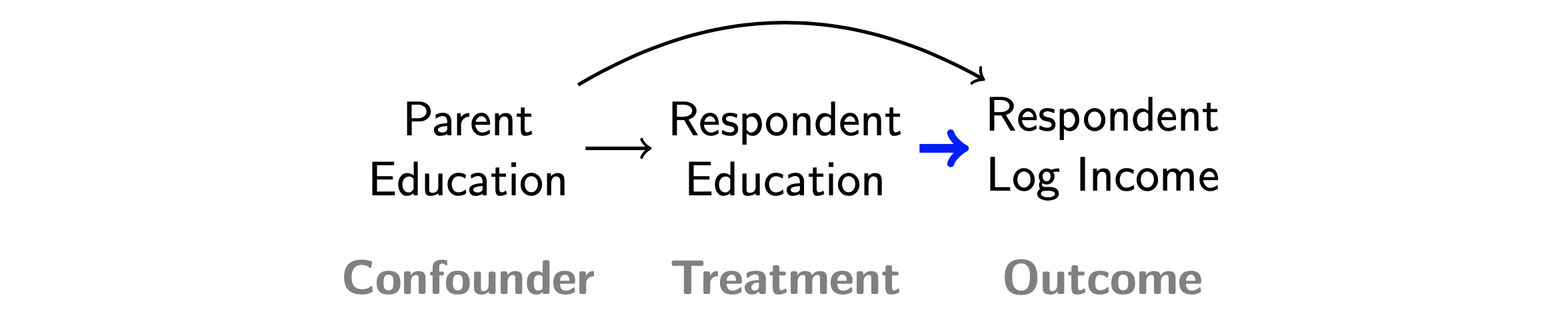
We illustrate by estimating the causal effect of education on income, where we assume that the causal effect is identified by statistical adjustment for parent education according to the DAG below.
Prepare: Load the data
First, prepare the environment and load data we used in the PSID Prediction Challenge.
library(tidyverse)
d <- read_csv("../psid_mobility_challenge/for_students/learning.csv") %>%
# Optional: Modify g2_educ so that it is an ordered factor
mutate(g2_educ = fct_relevel(g2_educ,
"Less than high school",
"High school",
"Some college",
"College")) %>%
# Optional: Rename variables to their roles for this lesson
rename(id = g3_id,
confounder = g2_educ,
treatment = g3_educ,
outcome = g3_log_income) %>%
select(id, confounder, treatment, outcome) %>%
print()
## # A tibble: 1,365 × 4
## id confounder treatment outcome
## <dbl> <fct> <chr> <dbl>
## 1 1 Less than high school High school 10.7
## 2 4 High school Some college 11.5
## 3 7 High school High school 11.2
## 4 9 Less than high school High school 10.7
## 5 10 Less than high school High school 10.7
## 6 11 High school Some college 11.3
## 7 12 High school Some college 11.5
## 8 13 Less than high school Less than high school 9.99
## 9 19 High school Less than high school 11.1
## 10 21 High school Less than high school 10.4
## # … with 1,355 more rows
Step 1. Create population subgroups and calculate conditional average effects
First, create population subgroups in which the confounders do not vary.
- within each subgroup, estimate the mean outcome in each treatment condition
- take the difference over treatment conditions of interest
This estimates conditional average causal effects: the average effect of a treatment on an outcome within population subgroups
conditional_average_effect <- d %>%
group_by(confounder, treatment) %>%
summarize(ybar = mean(outcome),
.groups = "drop") %>%
pivot_wider(names_from = treatment, values_from = ybar) %>%
# Focus on the effect of college vs high school
select(confounder, `College`, `High school`) %>%
mutate(effect_of_college = `College` - `High school`) %>%
print()
## # A tibble: 4 × 4
## confounder College `High school` effect_of_college
## <fct> <dbl> <dbl> <dbl>
## 1 Less than high school 11.5 10.8 0.740
## 2 High school 11.6 11.0 0.543
## 3 Some college 11.7 11.2 0.439
## 4 College 11.7 11.4 0.304
Step 2. Aggregate across subgroups
We might want a population claim: what is the average effect for everyone? An intervention to send respondents to college would not change the education of their parents. We will take the weighted average of conditional average causal effects, weighted by the size of each population subgroup.
First, calculate the size of each subgroup.
subpopulation_size <- d %>%
group_by(confounder) %>%
summarize(subgroup_size = n(),
.groups = "drop") %>%
print()
## # A tibble: 4 × 2
## confounder subgroup_size
## <fct> <int>
## 1 Less than high school 192
## 2 High school 664
## 3 Some college 296
## 4 College 213
Then, estimate the population average effect.
conditional_average_effect %>%
left_join(subpopulation_size, by = "confounder") %>%
summarize(population_average_effect = weighted.mean(effect_of_college,
w = subgroup_size)) %>%
print()
## # A tibble: 1 × 1
## population_average_effect
## <dbl>
## 1 0.511
Closing thoughts
Nonparametric adjustment illustrates a key concept: study the effect of a treatment by examining differences in outcomes across that treatment within subgroups defined by confounders.
In practice, there are often many confounders and nonparametric adjustment becomes impossible. The next page discusses parametric adjustment for those settings.